View on QuantumAI View on QuantumAI
|
 Run in Google Colab Run in Google Colab
|
 View source on GitHub View source on GitHub
|
|
try:
import cirq
except ImportError:
print("installing cirq...")
!pip install --quiet cirq
print("installed cirq.")
This notebook demonstrates how to use the functionality in cirq.experiments to run parallel XEB end-to-end. "Parallel" means we characterize multiple pairs simultaneously.
import cirq
import numpy as np
%matplotlib inline
from matplotlib import pyplot as plt
Parallel XEB with library functions
The entire XEB workflow can be run by calling cirq.experiments.parallel_two_qubit_xeb and the combined single-qubit randomized benchmarking (RB) and XEB workflows can be run by calling cirq.experiments.run_rb_and_xeb.
# Simulation
qubits = cirq.GridQubit.rect(3, 2, 4, 3)
result = cirq.experiments.parallel_two_qubit_xeb(
sampler=cirq.DensityMatrixSimulator(
noise=cirq.depolarize(5e-3), dtype=np.complex128
), # Any simulator or a ProcessorSampler.
qubits=qubits,
)
100%|██████████| 243/243 [02:45<00:00, 1.47it/s]
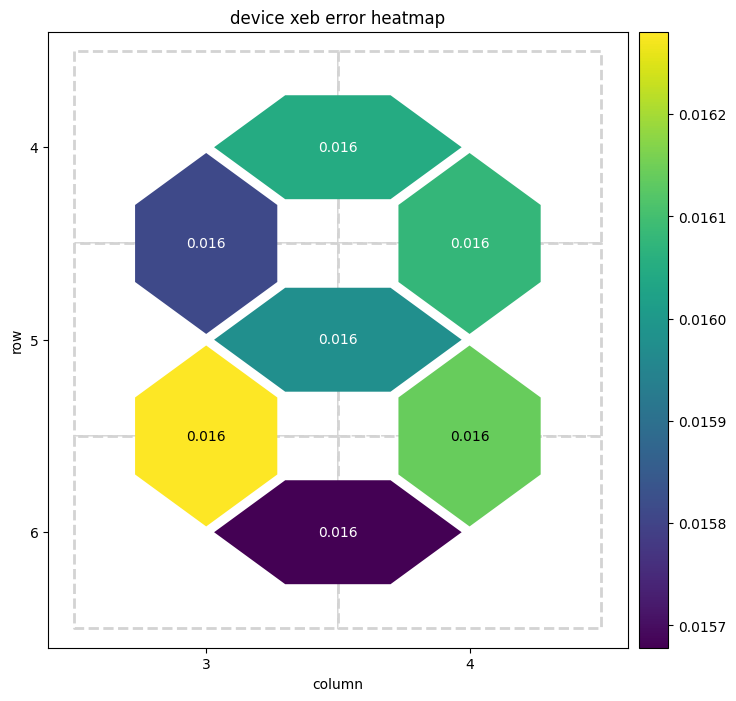
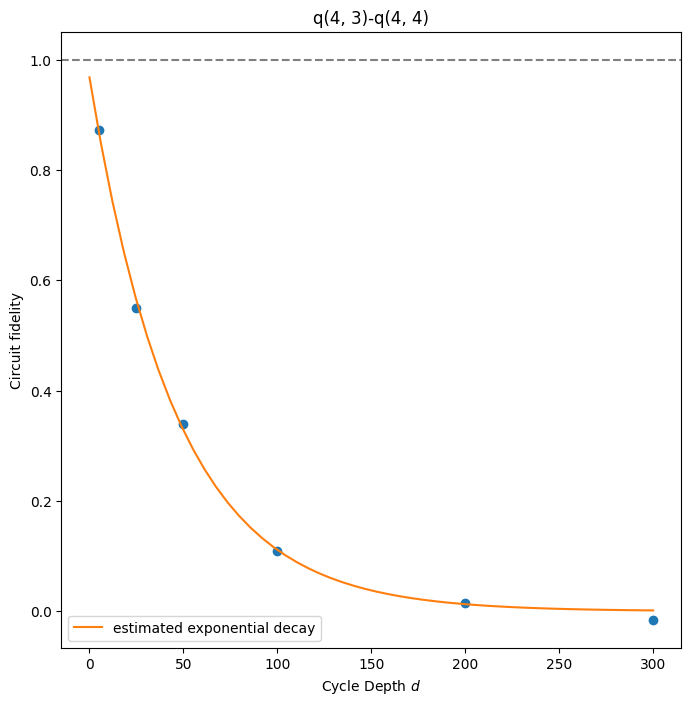
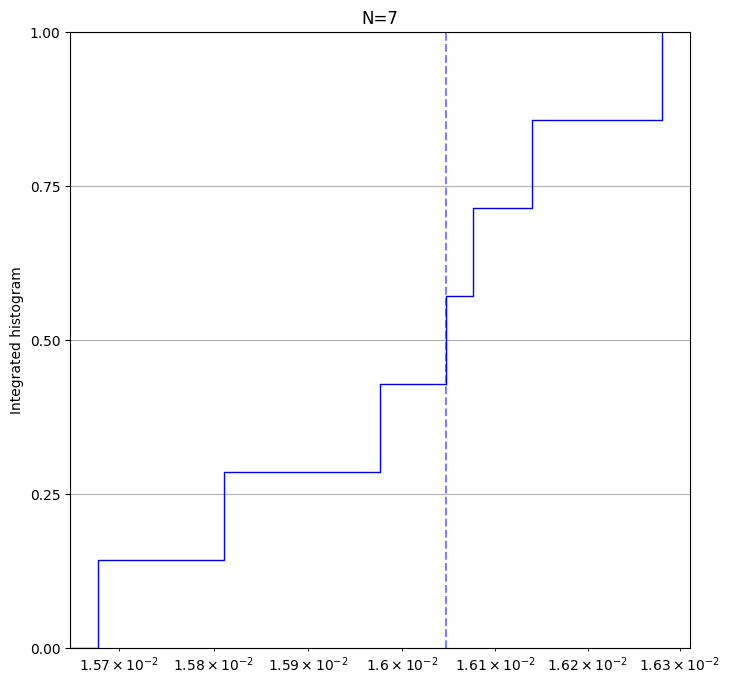
# The returned result is an instance of the `TwoQubitXEBResult` class which provides visualization methods like
result.plot_heatmap()
# plot the heatmap of XEB errors
result.plot_fitted_exponential(*qubits[:2])
# plot the fitted model of xeb error of a qubit pair.
result.plot_histogram(); # plot a histogram of all xeb errors.
# `TwoQubitXEBResult` also has methods to retrieve errors.
print('pauli errors:', result.pauli_error())
print('xeb errors:', result.xeb_error(*qubits[:2]))
print('xeb fidelity:', result.xeb_fidelity(*qubits[:2]))
pauli errors: {(cirq.GridQubit(4, 3), cirq.GridQubit(4, 4)): np.float64(0.019835896750389596), (cirq.GridQubit(4, 3), cirq.GridQubit(5, 3)): np.float64(0.019613922392582306), (cirq.GridQubit(4, 4), cirq.GridQubit(5, 4)): np.float64(0.020158079809996406), (cirq.GridQubit(5, 3), cirq.GridQubit(5, 4)): np.float64(0.019523058811304374), (cirq.GridQubit(5, 3), cirq.GridQubit(6, 3)): np.float64(0.019736423224559763), (cirq.GridQubit(5, 4), cirq.GridQubit(6, 4)): np.float64(0.019596043847401635), (cirq.GridQubit(6, 3), cirq.GridQubit(6, 4)): np.float64(0.019928119059041058)}
xeb errors: 0.015868717400311705
xeb fidelity: 0.9841312825996883
The run_rb_and_xeb method returns an object of type InferredXEBResult which is like TwoQubitXEBResult except that it removes the single-qubit errors obtained from the single-qubit randomized benchmarking (RB) experiment to isolate the error from the two qubit gate.
Step by step XEB
The rest of this notebook explains how the parallel_two_qubit_xeb works internally. Note that the notebook uses SQRT_ISWAP as the entangling gate while parallel_two_qubit_xeb and run_rb_and_xeb default to CZ.
Set up Random Circuits
We create a library of 10 random, two-qubit circuits using the sqrt(ISWAP) gate. These library circuits will be mixed-and-matched among all the pairs on the device we aim to characterize.
from cirq.experiments import random_quantum_circuit_generation as rqcg
circuit_library = rqcg.generate_library_of_2q_circuits(
n_library_circuits=20, two_qubit_gate=cirq.ISWAP**0.5, random_state=52
)
print(len(circuit_library))
20
# We will truncate to these lengths
max_depth = 100
cycle_depths = np.arange(3, max_depth, 20)
cycle_depths
array([ 3, 23, 43, 63, 83])
Determine the device topology
We will run on all pairs from a given device topology. Below, you can supply a device_name if you're authenticated to run on Google QCS. In that case, we will get the device object from the cloud endpoint and turn it into a graph of qubits. Otherwise, we mock a device graph by allocating arbitrary cirq.GridQubits to turn into a graph.
device_name = None # change me!
import cirq.contrib.routing as ccr
import networkx as nx
if device_name is None:
qubits = cirq.GridQubit.rect(3, 2, 4, 3)
# Delete one qubit from the rectangular arangement to
# 1) make it irregular 2) simplify simulation.
qubits = qubits[:-1]
sampler = cirq.DensityMatrixSimulator(noise=cirq.depolarize(5e-3))
graph = ccr.gridqubits_to_graph_device(qubits)
else:
import cirq_google as cg
sampler = cg.get_engine_sampler(device_name, gate_set_name='sqrt_iswap')
device = cg.get_engine_device(device_name)
qubits = sorted(device.qubits)
graph = ccr.gridqubits_to_graph_device(device.qubits)
pos = {q: (q.row, q.col) for q in qubits}
nx.draw_networkx(graph, pos=pos)
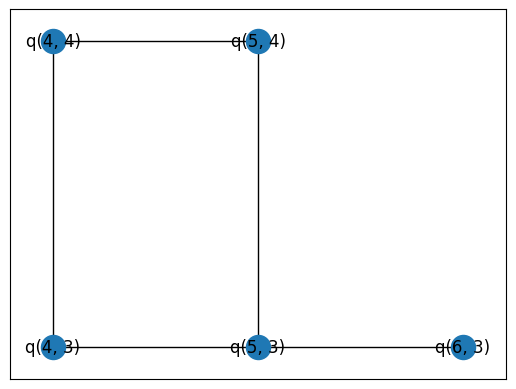
Set up our combinations
We take the library of two-qubit circuits in circuit_library and mix-and-match to sampled in parallel.
We will pass combs_by_layer and circuit_library to the sampling function which will "zip" the circuits according to these combinations. The outer list corresponds to the four cirq.GridInteractionLayers (one of four for the degree-four GridQubit-implied graph). The inner combinations matrix is a (n_combinations, n_pairs) ndarray of integers which index into the circuit library.
combs_by_layer = rqcg.get_random_combinations_for_device(
n_library_circuits=len(circuit_library), n_combinations=10, device_graph=graph, random_state=53
)
combs_by_layer
[CircuitLibraryCombination(layer=cirq.experiments.GridInteractionLayer(col_offset=0, vertical=True, stagger=True), combinations=array([[ 5, 16],
[12, 9],
[ 5, 18],
[11, 3],
[ 6, 9],
[13, 3],
[11, 6],
[14, 12],
[16, 10],
[18, 15]]), pairs=[(cirq.GridQubit(4, 4), cirq.GridQubit(5, 4)), (cirq.GridQubit(5, 3), cirq.GridQubit(6, 3))]),
CircuitLibraryCombination(layer=cirq.experiments.GridInteractionLayer(col_offset=1, vertical=True, stagger=True), combinations=array([[16],
[ 3],
[12],
[ 0],
[ 5],
[ 5],
[ 0],
[ 7],
[ 9],
[12]]), pairs=[(cirq.GridQubit(4, 3), cirq.GridQubit(5, 3))]),
CircuitLibraryCombination(layer=cirq.experiments.GridInteractionLayer(col_offset=1, vertical=False, stagger=True), combinations=array([[13],
[ 8],
[ 8],
[13],
[ 1],
[11],
[11],
[ 8],
[14],
[14]]), pairs=[(cirq.GridQubit(4, 3), cirq.GridQubit(4, 4))]),
CircuitLibraryCombination(layer=cirq.experiments.GridInteractionLayer(col_offset=0, vertical=False, stagger=True), combinations=array([[18],
[16],
[ 2],
[ 6],
[ 1],
[ 9],
[11],
[ 1],
[ 9],
[ 6]]), pairs=[(cirq.GridQubit(5, 3), cirq.GridQubit(5, 4))])]
Visualize
Here, we draw the four layers' active pairs.
fig, axes = plt.subplots(2, 2, figsize=(9, 6))
for comb_layer, ax in zip(combs_by_layer, axes.reshape(-1)):
active_qubits = np.array(comb_layer.pairs).reshape(-1)
colors = ['red' if q in active_qubits else 'blue' for q in graph.nodes]
nx.draw_networkx(graph, pos=pos, node_color=colors, ax=ax)
nx.draw_networkx_edges(
graph, pos=pos, edgelist=comb_layer.pairs, width=3, edge_color='red', ax=ax
)
plt.tight_layout()
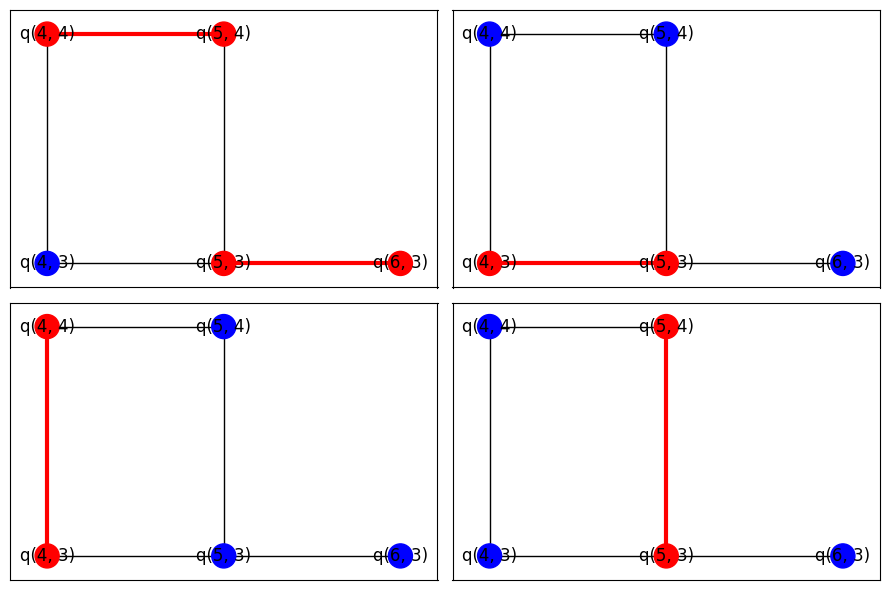
Take Data
The following call will execute the zipped circuits and sample bitstrings.
from cirq.experiments.xeb_sampling import sample_2q_xeb_circuits
sampled_df = sample_2q_xeb_circuits(
sampler=sampler,
circuits=circuit_library,
cycle_depths=cycle_depths,
combinations_by_layer=combs_by_layer,
shuffle=np.random.RandomState(52),
repetitions=10_000,
)
sampled_df
100%|██████████| 207/207 [00:45<00:00, 4.52it/s]
Benchmark Fidelities
from cirq.experiments.xeb_fitting import benchmark_2q_xeb_fidelities
fids = benchmark_2q_xeb_fidelities(
sampled_df=sampled_df, circuits=circuit_library, cycle_depths=cycle_depths
)
fids
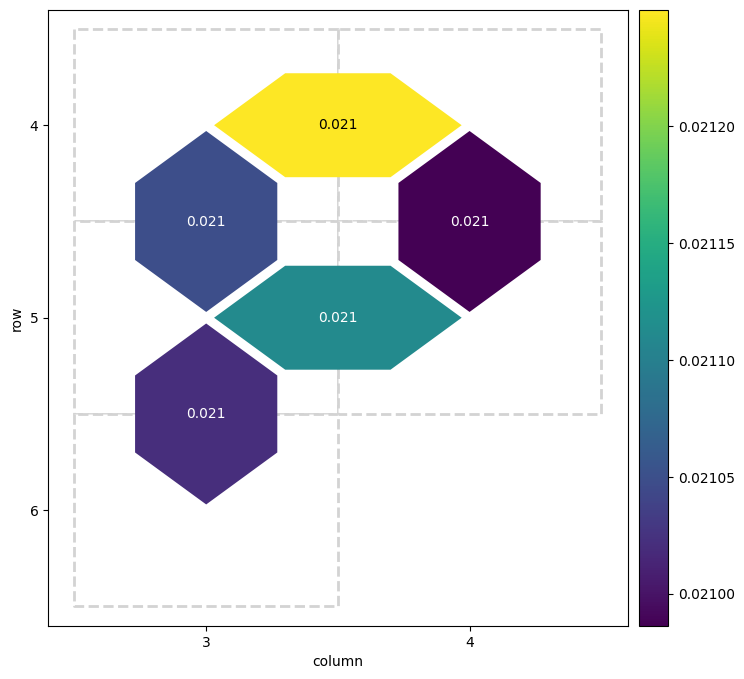
from cirq.experiments.xeb_fitting import fit_exponential_decays, exponential_decay
fidelities = fit_exponential_decays(fids)
heatmap_data = {}
for (_, _, pair), fidelity in fidelities.layer_fid.items():
heatmap_data[pair] = 1.0 - fidelity
cirq.TwoQubitInteractionHeatmap(heatmap_data).plot();
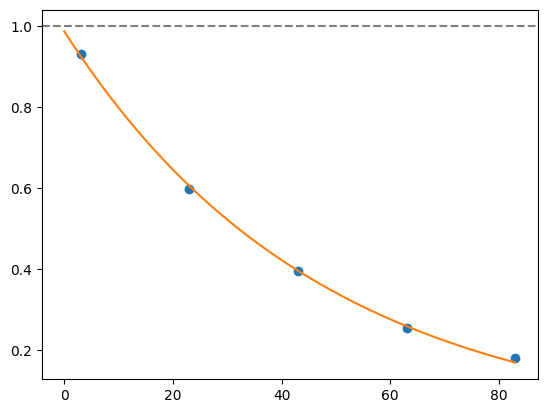
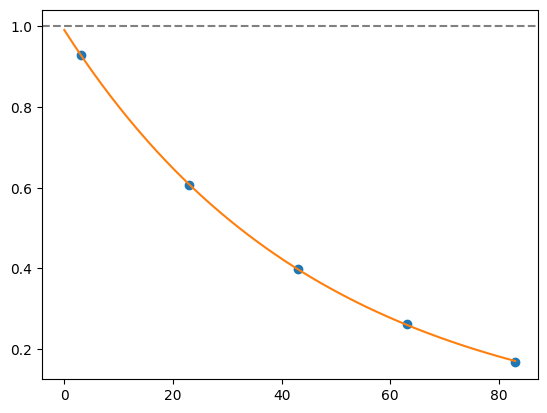
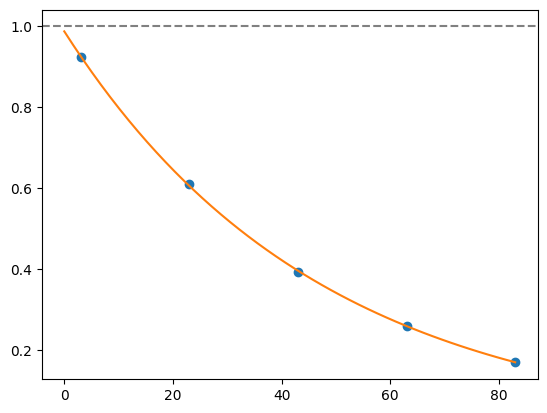
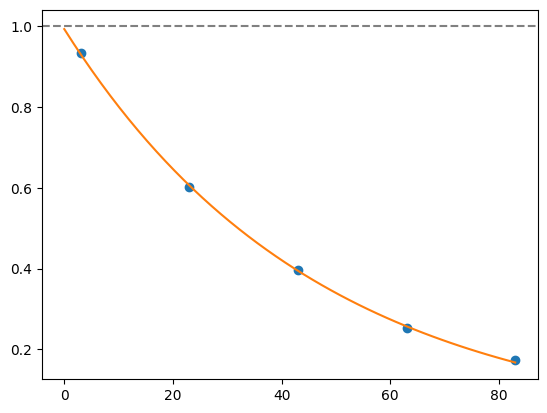
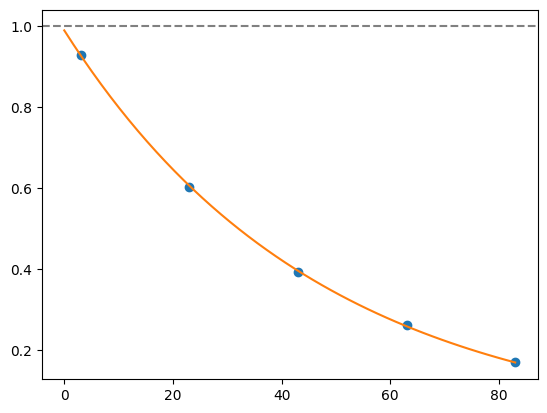
for i, record in fidelities.iterrows():
plt.axhline(1, color='grey', ls='--')
plt.plot(record['cycle_depths'], record['fidelities'], 'o')
xx = np.linspace(0, np.max(record['cycle_depths']))
plt.plot(xx, exponential_decay(xx, a=record['a'], layer_fid=record['layer_fid']))
plt.show()
import seaborn as sns
# Give each pair its own color
colors = sns.cubehelix_palette(n_colors=graph.number_of_edges())
colors = dict(zip(graph.edges, colors))
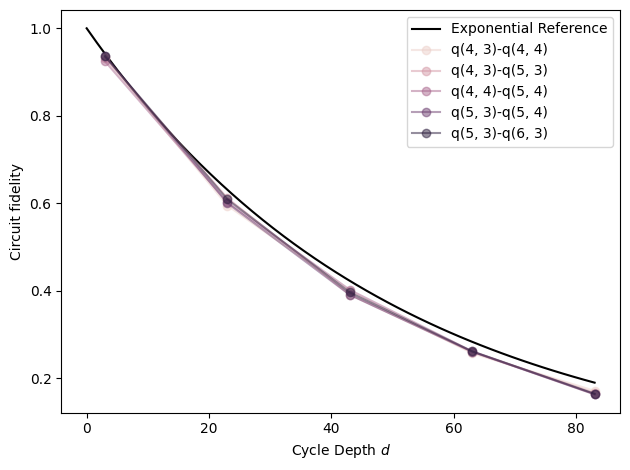
# Exponential reference
xx = np.linspace(0, fids['cycle_depth'].max())
plt.plot(xx, (1 - 5e-3) ** (4 * xx), label=r'Exponential Reference', color='black')
# Plot each pair
def _p(fids):
q0, q1 = fids.name
plt.plot(
fids['cycle_depth'],
fids['fidelity'],
'o-',
label=f'{q0}-{q1}',
color=colors[fids.name],
alpha=0.5,
)
fids.groupby('pair').apply(_p)
plt.ylabel('Circuit fidelity')
plt.xlabel('Cycle Depth $d$')
plt.legend(loc='best')
plt.tight_layout()
/tmpfs/tmp/ipykernel_78850/3110110171.py:25: DeprecationWarning: DataFrameGroupBy.apply operated on the grouping columns. This behavior is deprecated, and in a future version of pandas the grouping columns will be excluded from the operation. Either pass `include_groups=False` to exclude the groupings or explicitly select the grouping columns after groupby to silence this warning.
fids.groupby('pair').apply(_p)
Optimize PhasedFSimGate parameters
We know what circuits we requested, and in this simulated example, we know what coherent error has happened. But in a real experiment, there is likely unknown coherent error that you would like to characterize. Therefore, we make the five angles in PhasedFSimGate free parameters and use a classical optimizer to find which set of parameters best describes the data we collected from the noisy simulator (or device, if this was a real experiment).
import multiprocessing
pool = multiprocessing.get_context('spawn').Pool()
from cirq.experiments.xeb_fitting import (
parameterize_circuit,
characterize_phased_fsim_parameters_with_xeb_by_pair,
SqrtISwapXEBOptions,
)
# Set which angles we want to characterize (all)
options = SqrtISwapXEBOptions(
characterize_theta=True,
characterize_zeta=True,
characterize_chi=True,
characterize_gamma=True,
characterize_phi=True,
)
# Parameterize the sqrt(iswap)s in our circuit library
pcircuits = [parameterize_circuit(circuit, options) for circuit in circuit_library]
# Run the characterization loop
characterization_result = characterize_phased_fsim_parameters_with_xeb_by_pair(
sampled_df,
pcircuits,
cycle_depths,
options,
pool=pool,
# ease tolerance so it converges faster:
fatol=1e-2,
xatol=1e-2,
)
characterization_result.final_params
{(cirq.GridQubit(5, 3),
cirq.GridQubit(5, 4)): {'theta': np.float64(-0.8367987404054755), 'zeta': np.float64(0.10041558244274296), 'chi': np.float64(-0.07510134699446311), 'gamma': np.float64(-0.017183029157944202), 'phi': np.float64(0.004193753360909505)},
(cirq.GridQubit(4, 3),
cirq.GridQubit(4, 4)): {'theta': np.float64(-0.8474371212531532), 'zeta': np.float64(-0.10035248634877036), 'chi': np.float64(-0.07034612478660182), 'gamma': np.float64(0.050899749487735915), 'phi': np.float64(-0.041682247860173494)},
(cirq.GridQubit(4, 3),
cirq.GridQubit(5, 3)): {'theta': np.float64(-0.7552448278453846), 'zeta': np.float64(0.033253969974588855), 'chi': np.float64(-0.03479666867301002), 'gamma': np.float64(0.03136281359839602), 'phi': np.float64(-0.06806601866053645)},
(cirq.GridQubit(4, 4),
cirq.GridQubit(5, 4)): {'theta': np.float64(-0.7790601994013041), 'zeta': np.float64(0.032579693881346745), 'chi': np.float64(-0.02279511443835067), 'gamma': np.float64(0.00337801077356593), 'phi': np.float64(0.0352923155799907)},
(cirq.GridQubit(5, 3),
cirq.GridQubit(6, 3)): {'theta': np.float64(-0.758190997670132), 'zeta': np.float64(0.004304546463840975), 'chi': np.float64(-0.049149081691340135), 'gamma': np.float64(0.0020049245053540667), 'phi': np.float64(0.025945488060518777)} }
characterization_result.fidelities_df
from cirq.experiments.xeb_fitting import before_and_after_characterization
before_after_df = before_and_after_characterization(fids, characterization_result)
before_after_df
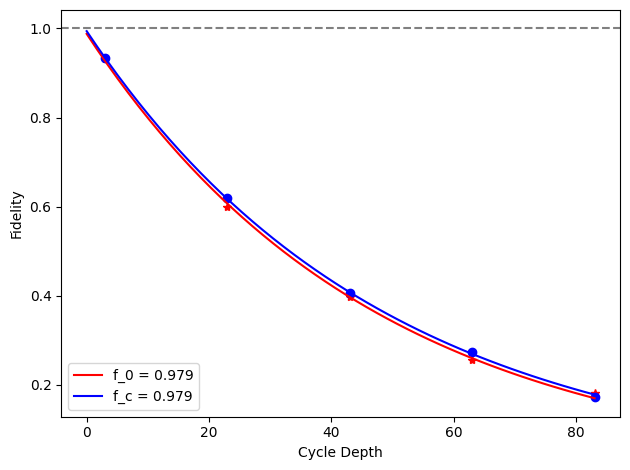
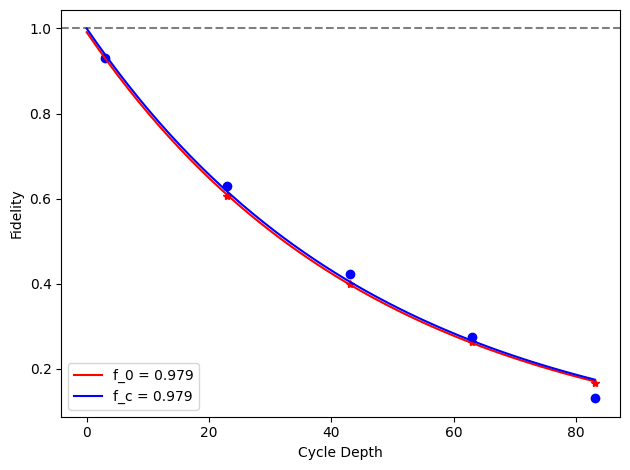
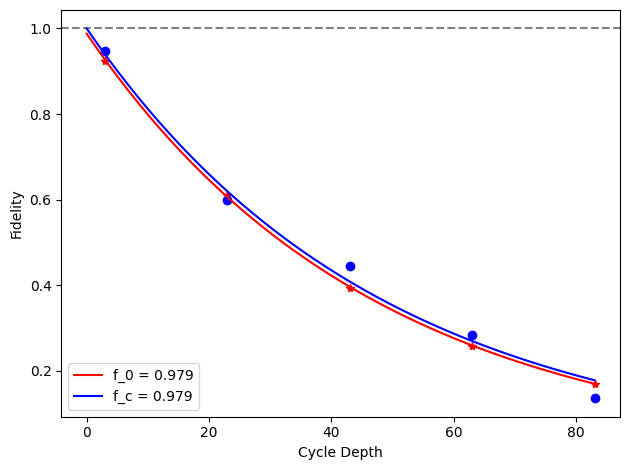
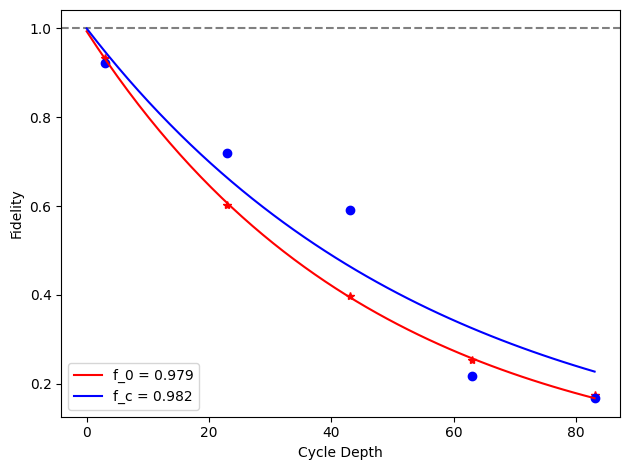
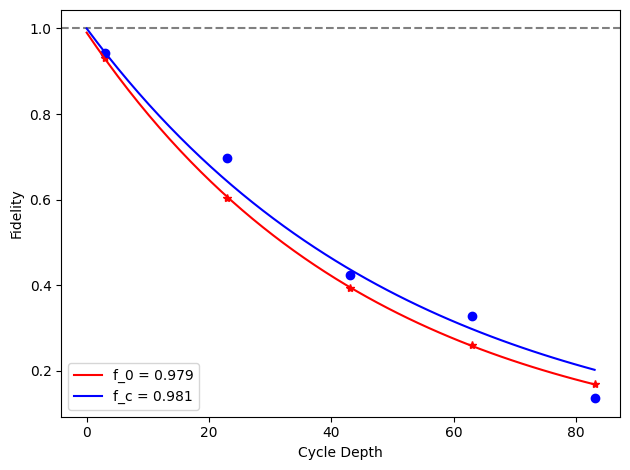
for i, row in before_after_df.iterrows():
plt.axhline(1, color='grey', ls='--')
plt.plot(row['cycle_depths_0'], row['fidelities_0'], '*', color='red')
plt.plot(row['cycle_depths_c'], row['fidelities_c'], 'o', color='blue')
xx = np.linspace(0, np.max(row['cycle_depths_0']))
plt.plot(
xx,
exponential_decay(xx, a=row['a_0'], layer_fid=row['layer_fid_0']),
color='red',
label=f'f_0 = {row["layer_fid_0"]:.3f}',
)
plt.plot(
xx,
exponential_decay(xx, a=row['a_c'], layer_fid=row['layer_fid_c']),
color='blue',
label=f'f_c = {row["layer_fid_c"]:.3f}',
)
plt.xlabel('Cycle Depth')
plt.ylabel('Fidelity')
plt.legend(loc='best')
plt.tight_layout()
plt.show()